Computational Models
Special considerations to prepare metadata files for Computational Datasets
This page provides information on how to compile the code_description.xlsx table. In addition to the links, descriptions, helpers and comments embedded into the Excel file, this documentation should help resolve doubts - if you have further questions, feel free to reach out to [email protected].
This documentation is meant for submission of SPARC computational datasets that were not generated or onboarded on o²S²PARC. If your dataset is on o²S²PARC, code_description.xlsx can be generated automatically and downloaded, based on information provided through its GUI (see o²S²PARC documentation for details).
To illustrate and clarify the guidance in this document, an example code_description.xlsx file is provided here.
Which information is mandatory?
The cells background colors demark the following:
Blue: mandatory for every computational model publication on the Portal, and the “Link” column (preferred) or the “Text” column must be filled in
Green: everyone should try to fill these out, if possible
Yellow: filling out these fields is voluntary and they may in some instances not even be relevant or required for downstream consumers
Gray: these cells are not meant to be filled out
Table structure
The requested information is organized in three blocks:
- Ontological Terms and Identifiers (Medatada element, rows 1-5)
- Ten Simple Rules (TRSs) rating rubric (rows 7-25)
- Input/Output information (rows 27-39)
Ontological Terms and Identifiers
Research Resource Identifiers (#RRID) are ID numbers assigned to help researchers cite key resources (antibodies, model organisms and software projects) in the biomedical literature to improve transparency of research methods. As part of the cMIS, such RRIDs can be provided to unambiguously specify employed computational tools and software (provided that associated RRIDs exist for the employed software).
RRID Term: name of software, tool or other computational resources used in your project. You can provide zero, one, or multiple terms.
RRID Identifier: RRID identifier associated with the RRID Term in the row above (you can find them on SciCrunch). If no RRID is available, leave it empty or specify ‘N/A’.
Ontology Terms are standardized terms describing biological entities (e.g. organ, system, species,...) for the purpose of reducing ambiguity and facilitating harmonization and search. SPARC ontologies can be queried on SciCrunch.
Ontology Term: ontological terms describing biological entities (e.g. organ, system, species,...).You can provide zero, one or multiple terms.
Ontology Identifier: ontological identifiers associated with the Ontology Term in the row above (you can find them on SciCrunch).
Note: RRID Terms/Identifiers are not related to Ontology Term/Identifier, so rows 2+3 and 4+5 should be read and/or filled out independently.
Ten Simple Rules (TRSs) rating rubric
The TSR is a communication tool for modelers to organize their model development process and present it coherently to each stakeholder's interests (see 10 Simple Rules with Conformance Rubric). The TSR was elaborated by the Committee on Credible Practice of Modeling & Simulation in Healthcare. It foresees that model creators both self-rate their level of adherence to these ten rules, as well as formulate a goal of the level of adherence that they aim for. For the cMIS (and o²S²PARC) the original TSR were slightly adapted in accordance to specific o²S²PARC needs and requirements from the SPARC Knowledge Management core. In addition to the ratings, TSR relevant documentation and/or support for the rating choice should be provided (as free-form text, or links to, e.g., associated papers or repositories).
TSR1, TSR4 and TSR6 are mandatory, so you should provide a rating level (choose it from the drop-down menu) and at least one link or text as demonstration or explanation.
The TSRs rating rubric refers to the entire modeling package submitted. As such only one rating should be provided for each rule (and not multiple ones corresponding to each RRID/Ontology Term).
In accordance with the 10 Simple Rules with Conformance Rubric document referenced above, the rating levels for the TSRs are defined as:
- Comprehensive:
- Can be understood by non-modeling/simulation practitioners familiar with the application domain and the intended context of use.
- Outreach capability: outreach to application-domain experts who may not be modeling and simulation experts.
- Extensive:
- Can be understood by modeling/simulation practitioners not familiar with the application domain and the intended context of use.
- Outreach capability: outreach to modeling and simulation practitioners who may not be application-domain experts.
- Adequate:
- Can be understood by modeling/simulation practitioners familiar with the application domain and the intended context of use.
- Outreach capability: outreach to application-domain specific modeling and simulation practitioners.
- Partial:
- Unclear to the modeling/simulation practitioners familiar with the application domain and the intended context of use.
- Outreach capability: outreach to application-domain specific modeling and simulation practitioners.
- Insufficient:
- Missing or greatly incomplete information to properly evaluate the conformance with the rule.
- Outreach capability: none or very limited.
The rating levels described above are also summarized in Table 1:
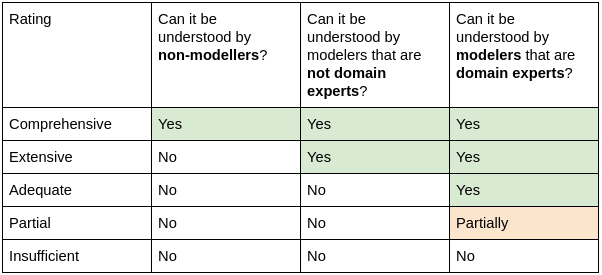
Table 1: Simplified summary of the 10 Simple Rules conformance levels.
For all the other TSRs if you provide a rating, you also have to provide a link or text as support. If you wish, you can provide more than one link and you can do that by adding new columns (e.g. at the right of Column D).
For target justification, links to published manuscripts are valid. For example, this is typically the case for “TSR4: Explicitly Listed Limitations” and other rules.
o²S²PARC provides a dedicated dialog to display and fill in the TSR-related information. This information can then be exported to generate a downloadable cMIS file on o²S²PARC.
Input/Output Information
Proper definition of input and output ports provides clarity about the required and acceptable inputs (e.g., type, dimensions, units, acceptable range). On o²S²PARC, it also forms the basis for parameterized modeling, for determining compatibility of data (e.g., from the Portal or from an upstream service), for automatically converting between units, and for proposing compatible analysis/modeling workflows for data on the Portal (and at a later point for file-level search/filtering on the Portal for suitable model input data).
You should provide the Number of Inputs (cell 27D) and the Number of Outputs (cell 34D) for your computational dataset. Then, for each input (output) you should complete a column - one column per input (output) - and report corresponding properties in the respective rows. Properties include Name, Type, human-readable Description, Units, Default Value and Constraints (if applicable). For example, if the computational dataset has 2 inputs, the first input will be specified in rows 28-33, column D; the second input in rows 28-33, column E.
Updated 9 months ago