Submission Overview
What is the process to publish Data & Models on the SPARC Portal?
SPARC is an open data repository platform specializing in hosting and publishing multi-modal and FAIR data on the peripheral nervous system and its CNS-end organ interactions. For that reason, we work with those publishing items on the Portal to make sure they publish a product that is usable to others-although many find their published items are easy to use for their own internal needs as well.
Before getting started, please ensure your request has been approved to ensure this is a good home for your data or model. Find out more at https://sparc.science/share-data.
General Requirements
SPARC upholds high standards for publishing datasets and models on its Portal, ensuring accessibility, structure, and AI readiness. Our expert Curation team provides hands-on guidance to help investigators create well-structured datasets with a dedicated landing page, DOI, and linked protocols.
To support collaboration and data reuse, SPARC follows consistent standards that enhance browsing, searching, and dataset comprehension:
- Datasets must adhere to the SPARC Data Structure (SDS) for standardized file organization and naming conventions.
- Each submission must include a detailed, experimental protocol, with protocols.io being the recommended platform.
- SPARC accepts specific file formats—investigators should reach out to the Curation team if their format is not listed.
Certain funding programs may have additional requirements, and we are here to help researchers navigate them. With structured metadata and expert guidance, SPARC ensures that published datasets are well-curated for reusability, facilitating future discoveries and valuable scientific insights.
Overview of the Curation and Publication Process
The figure below gives an overview of the full process. Additional documentation is shared in the rest of this section.
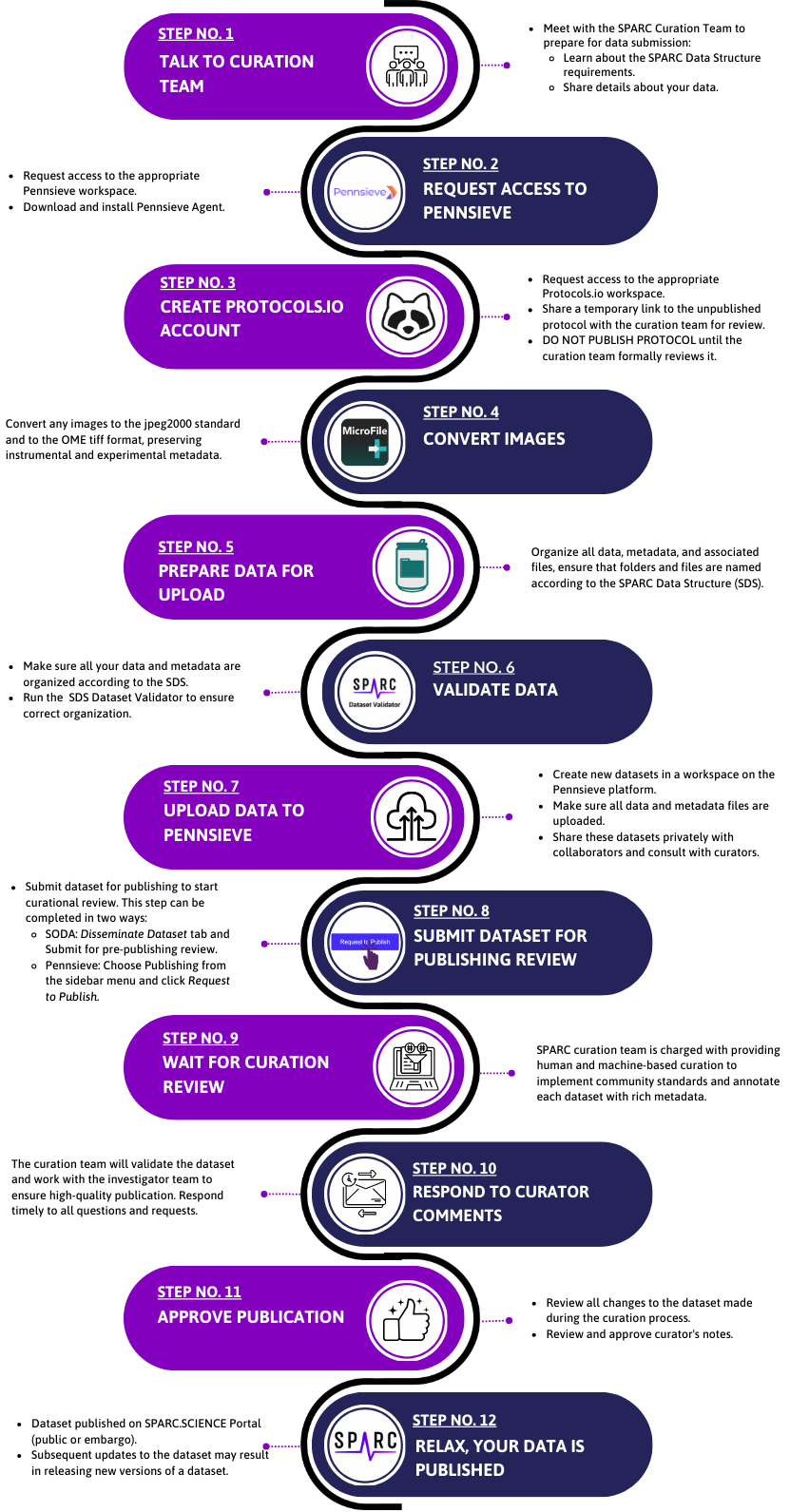
Dataset Publishing Process Expectations
Think of publishing a dataset as a process similar to publishing a manuscript. Preparing datasets and protocols for submission can take quite a bit of time, especially for those submitting for the first time.
Please keep in mind that the Curation team manages multiple dataset submissions simultaneously:
- Once submitted, the Curation team will usually provide initial feedback to you in ~7 to 10 days
- At this point you will iterate on corrections and improvements. This time depends on how well the data or protocol is prepared and responsiveness of investigators.
- Curation team tries to give a rapid turnaround on feedback to modifications
Once the Curation team performs final checks and proofs your dataset for publication, you, as the investigator and dataset owner, must sign off on the changes before publication.
Datasets are published on Fridays:
- The dataset will be visible on the SPARC Portal
- The DOI will resolve to the SPARC Portal URL
- Search may not work for a few days
Expect to allocate a minimum of one month (two is better!) to complete the full process:
Timeline Expectations
- The first time you prepare a dataset, it can take a while - over time, it gets faster and easier!
- Think of this as a process similar to publishing a manuscript; preparing the manuscript, submitting it for review and revising can take several weeks. Plan on a minimum of a month for the process, more is better.
- The curation team is your reviewer; the platform is your publisher. Just as with an article, there are standards, quality checks and processes that must be performed.
When you start preparing your manuscript, start preparing your dataset
Updated 8 months ago