Integrated Maps Viewer
What is the Integrated Maps Viewer?
The Integrated Map Viewer provides the point of entry to the different AC flatmaps.
What can I do with the Integrated Maps Viewer?
The integrated maps viewer can be used to access and interact with:
- Flatmaps
- 3D maps
- Anatomical scaffolds
- Data
- Simulations
Access the integrated maps viewer
Learn more about maps and how to access them in the SPARC App's Maps section.
The Display
Flatmap Viewer
The Integrated Maps Viewer's primary interface is the AC Flatmap Viewer. Learn more about how to use it in the AC Flatmap section.
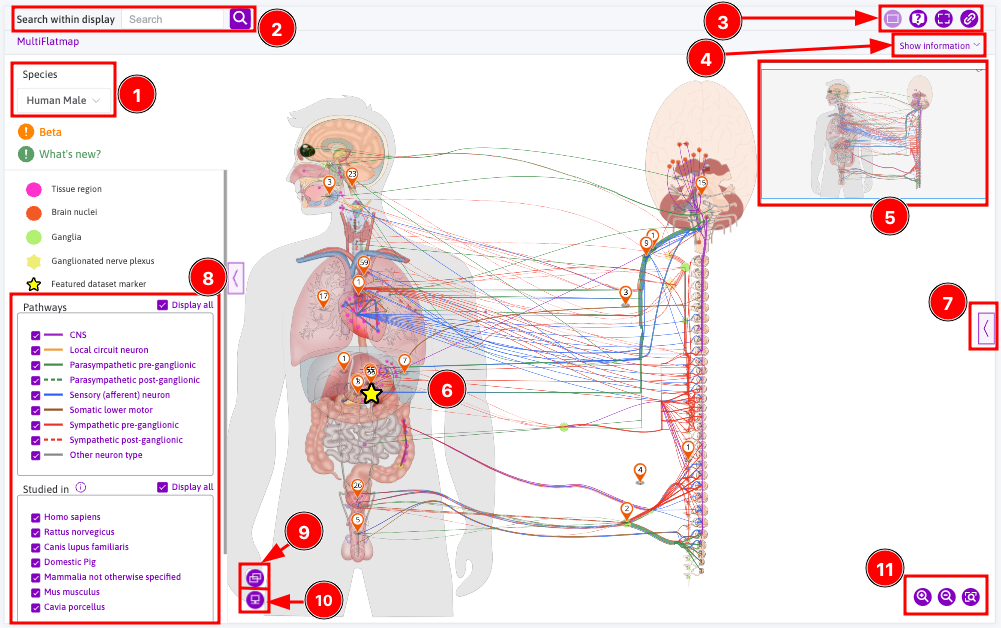
sidebar
What can I do with the integrated maps viewer?
Interact with flatmaps
You can interact with the flatmap itself (Figure 1, 6). learn how to xxx
Open 3D maps
3D maps can be opened from the Open new map tooltip.
Learn how to use 3D Maps in detail
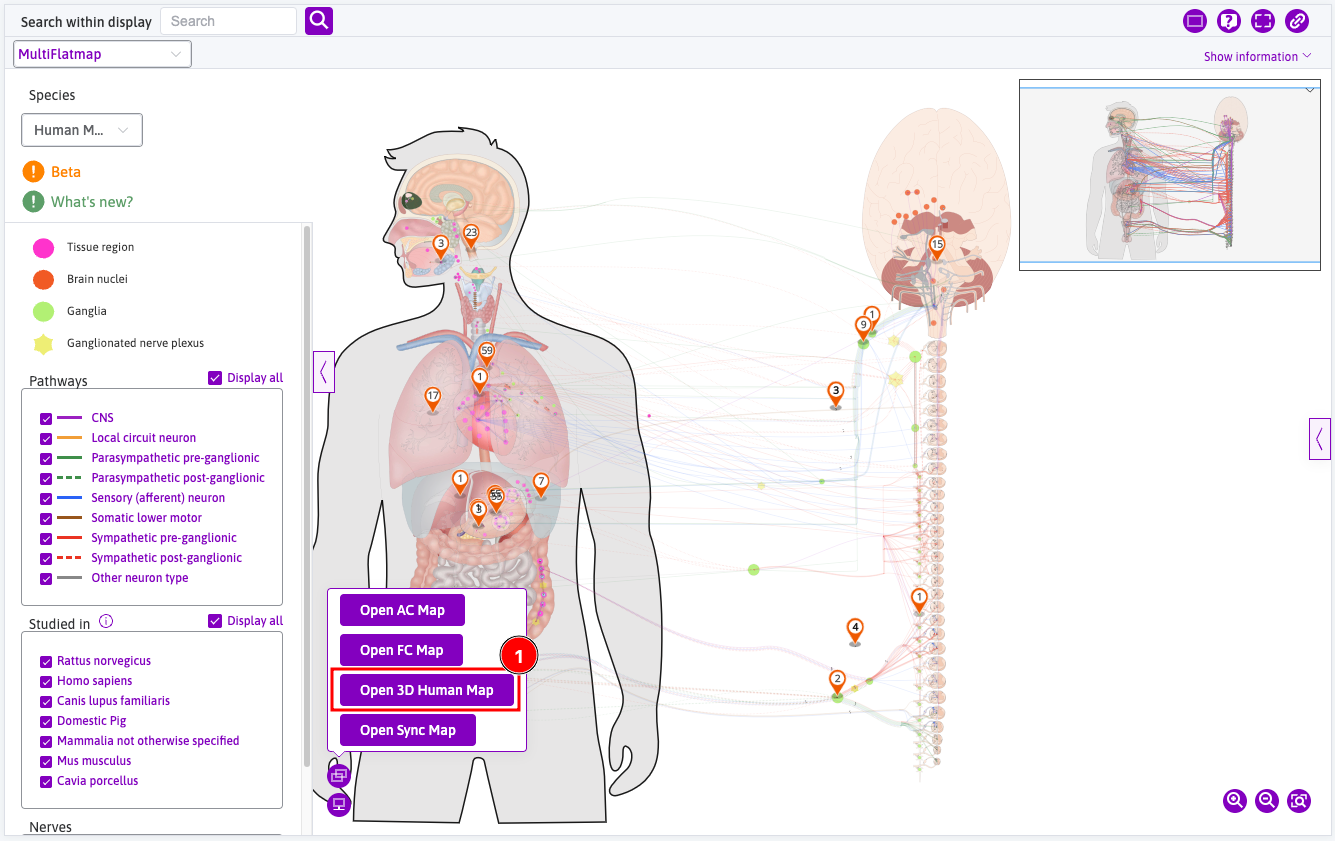
Anatomical Models
Access to anatomical scaffolds can be done either through the flatmap itself or through the sidebar, as illustrated here.
Simulations
Access to simulations is done through the sidebar, as illustrated here.
Access Data & Models
Datasets views
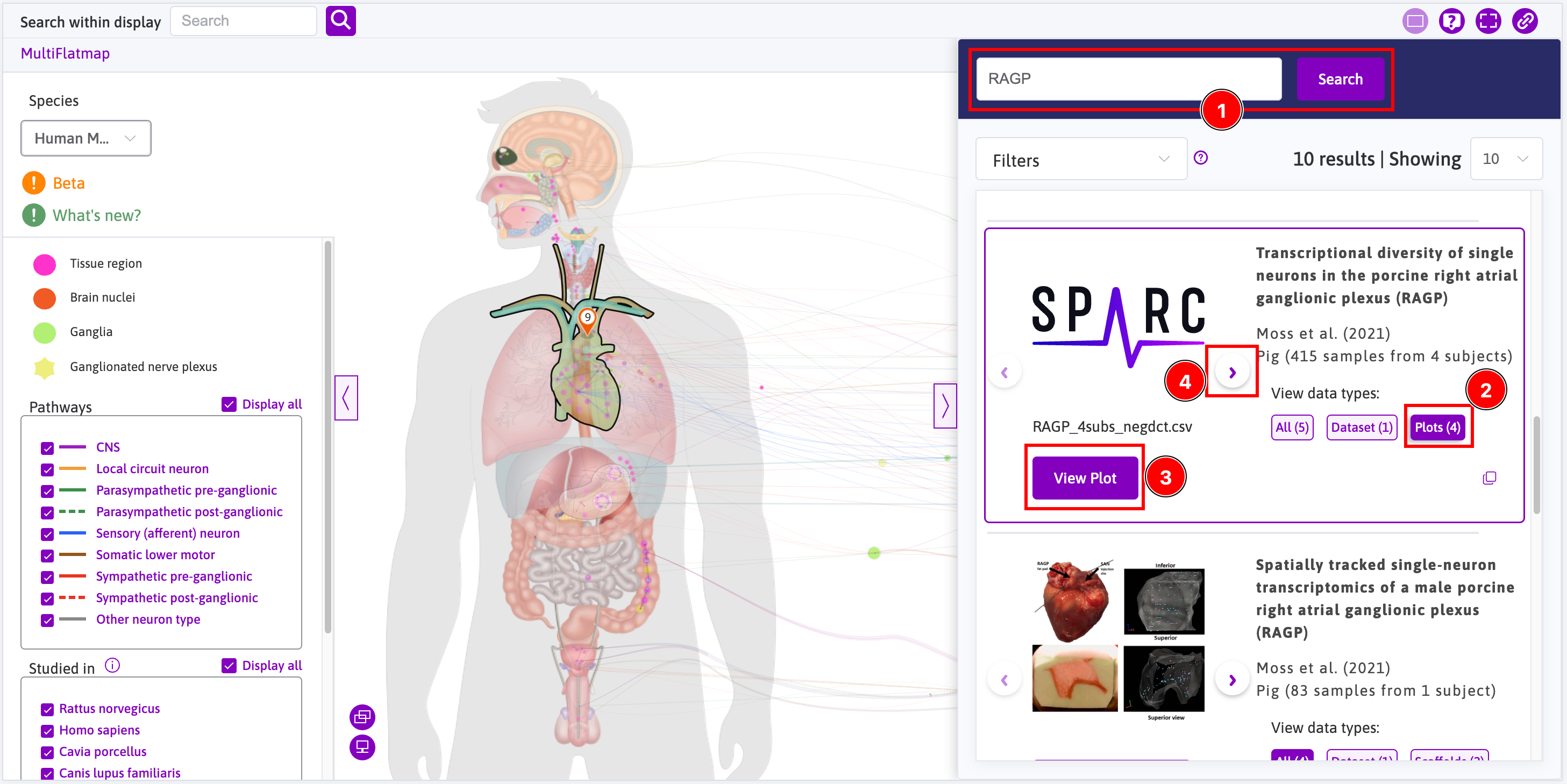
Access to data is done through the sidebar where you can search for datasets that contain data that can be rendered using the Data viewer. For instance, open the sidebar and:
- Search for
RAGP(i.e. right atrial ganglionic plexus); - Click on the
Plots (4)button; - Click on the
View Plotbutton; - Click on the right arrow button and go back to step 3 until you have opened all four plots;
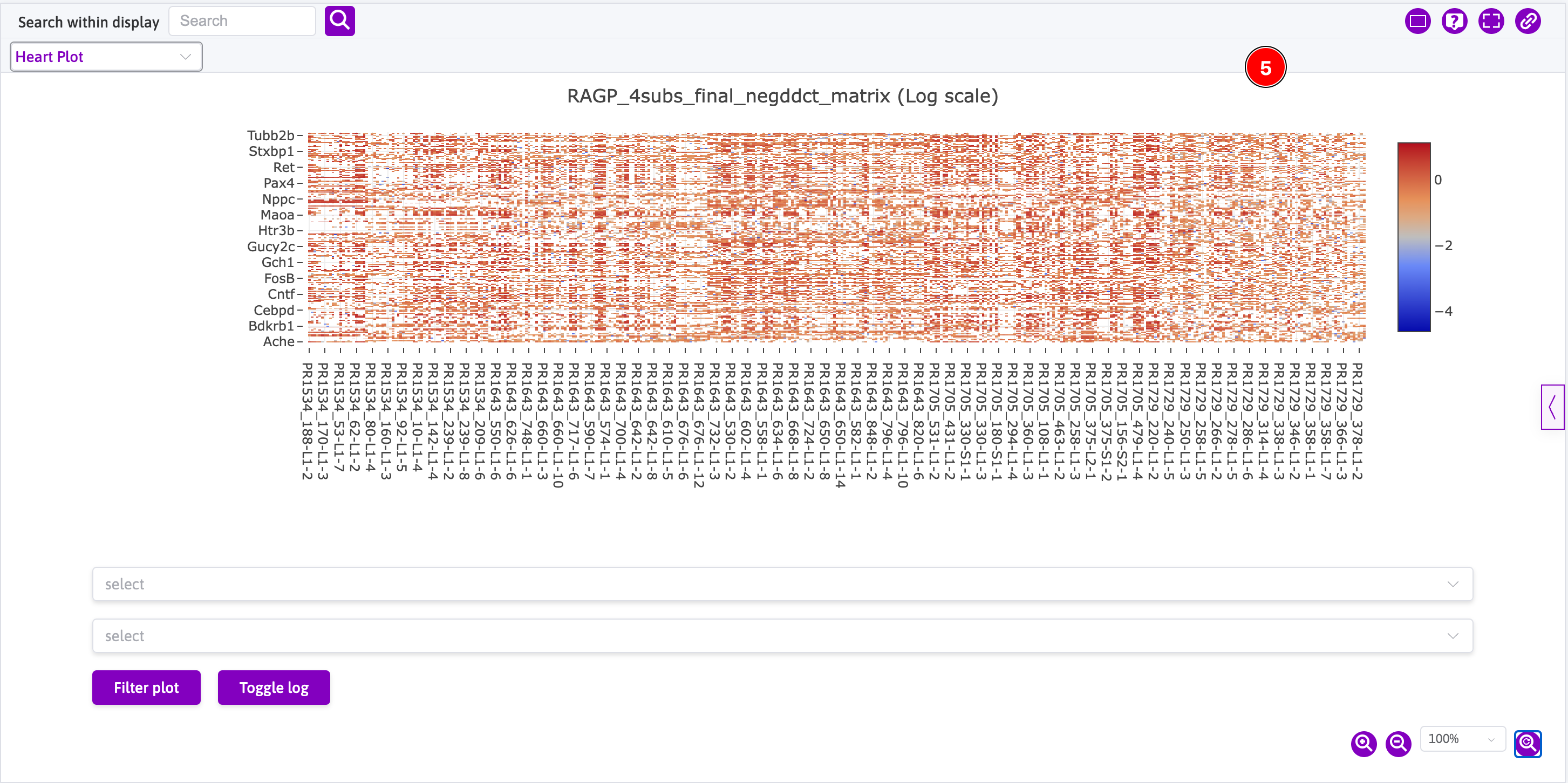
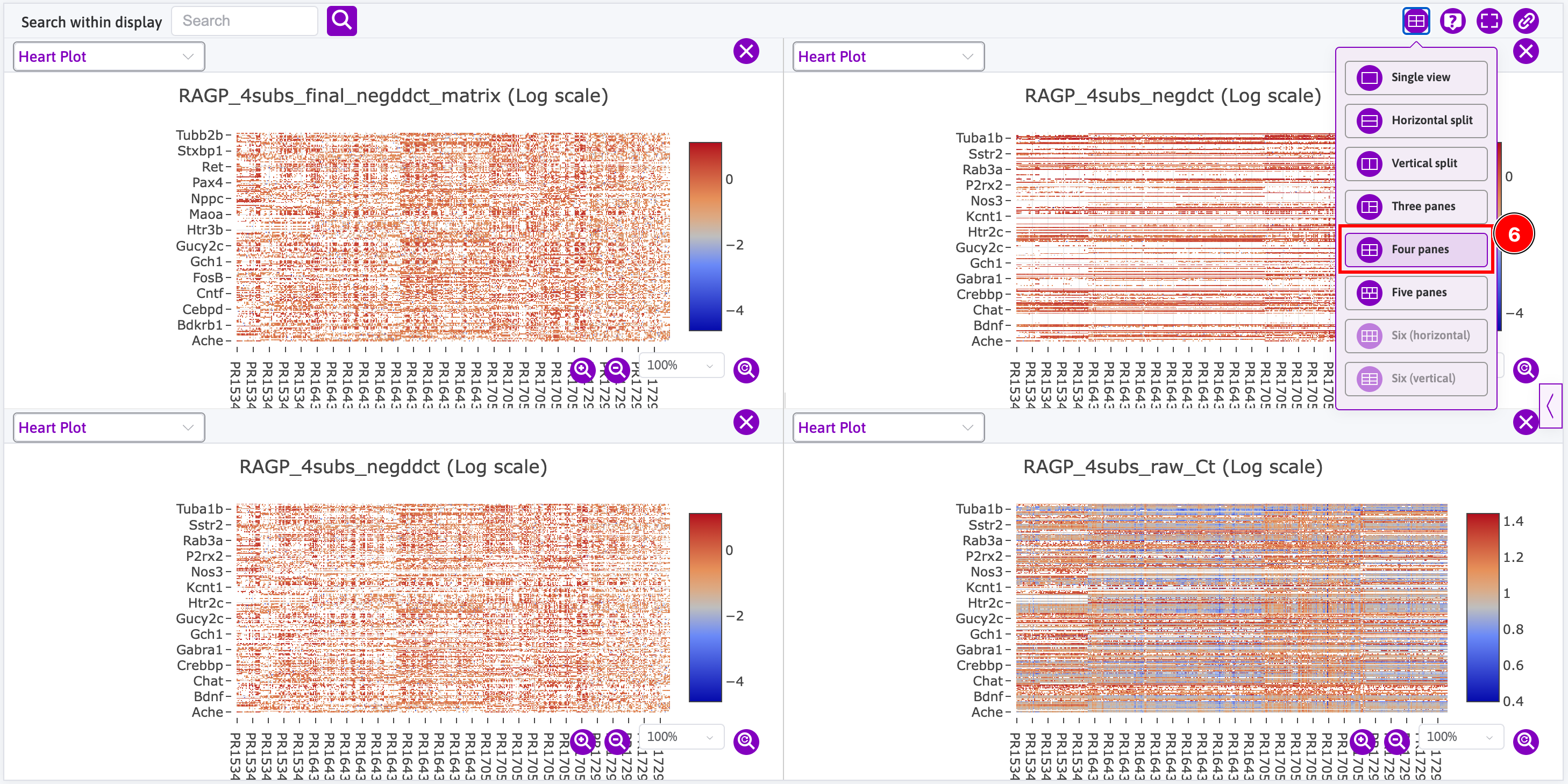
- This is how one plot is initially rendered; and
- This is how all four plots are eventually rendered using the 4-page view.
Computational Models and Simulations
Access to simulations is done through the sidebar, as illustrated here.
Explore Connectivity
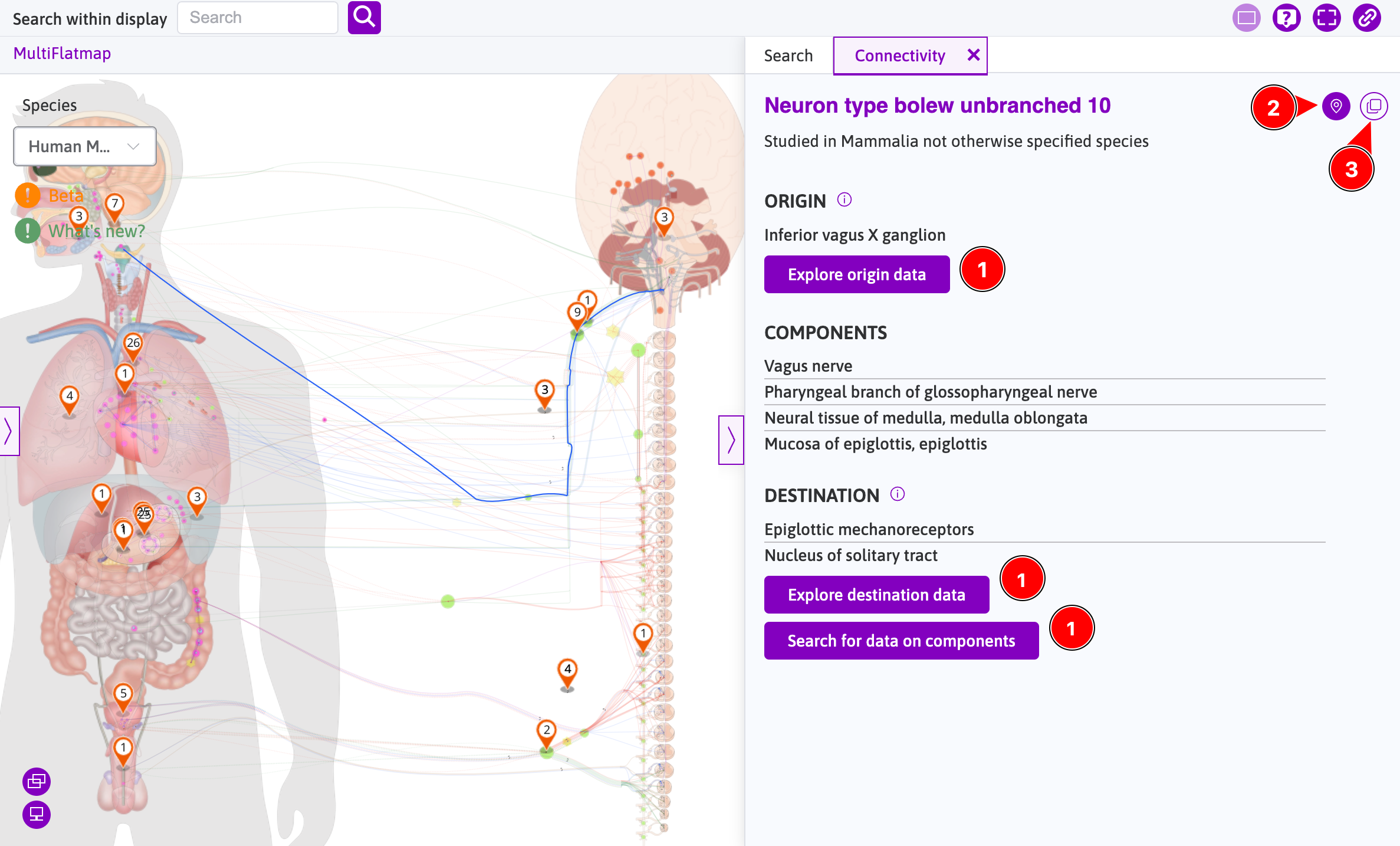
When the Connectivity tab is opened in the sidebar by clicking a neuron on flatmap (e.g., clicking "Neuron type bolew unbranched 10"), the information in the connectivity tab is connected with the sidebar's Search tab and also with the flatmap.
- Clicking
Explore origin data,Explore destination data, orSearch for data on componentswill switch to theSearchtab by applying the connectivity data in filters. - Clicking
Show connectivity on mapwill move the map to show the highlighted area. - Clicking
Copy to clipboardbutton will copy the connectivity information to clipboard.
Known Issues
Limit on available WebGL contexts
Certain data viewers on the SPARC Portal (i.e., scaffold and flatmap viewers) rely on specialized WebGL contexts to render demanding graphics. Browsers and systems impose a limit on the number of active contexts. Once this limit is reached, the browser typically discards the oldest context to fulfill the newest request. Map-Viewer attempts to manage this process behind the scenes. In situations where automatic handling could cause further display errors, a button will be provided for you to restore the display manually.
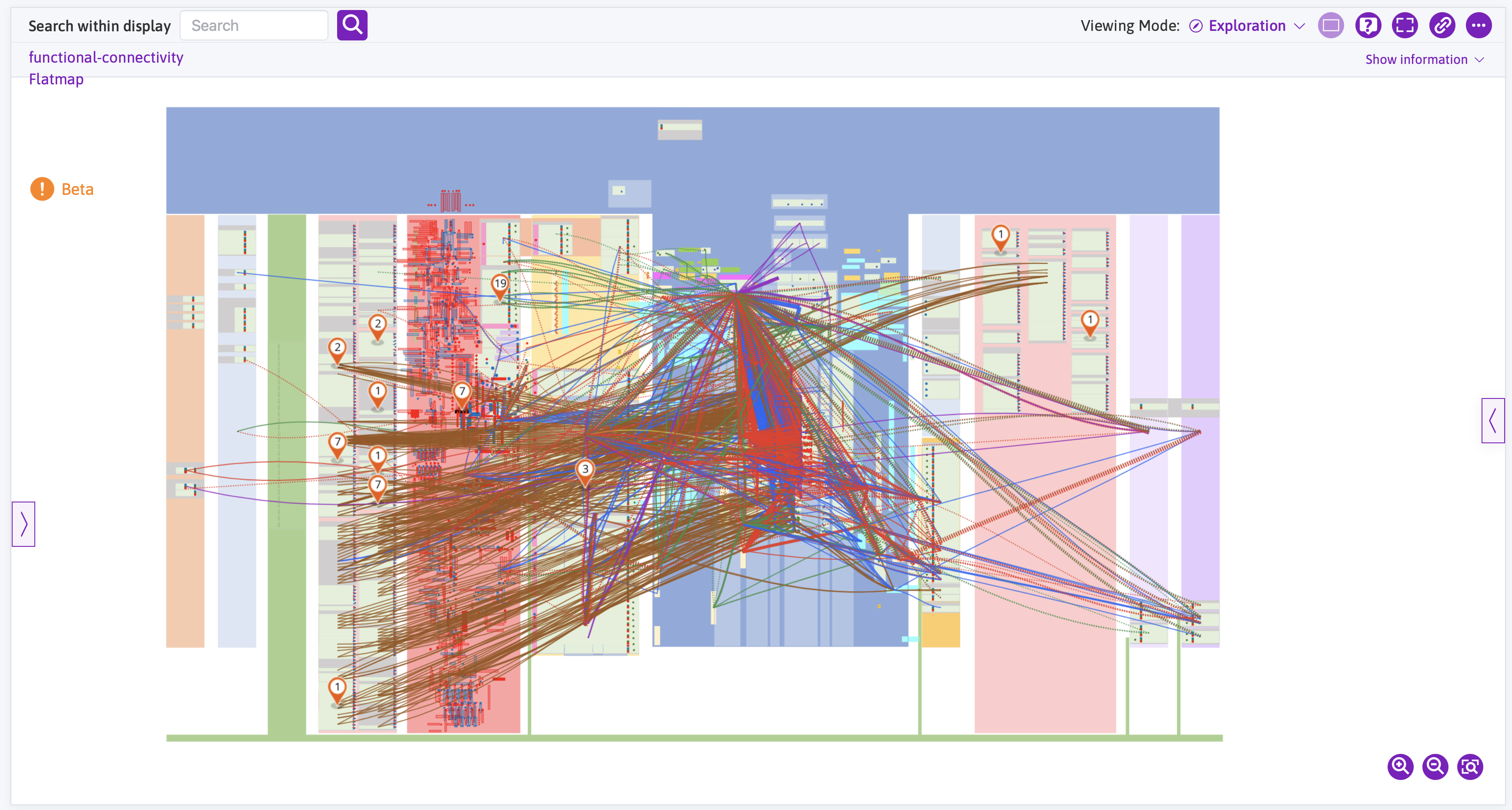
Viewing the legacy FC map
The legacy functional connectivity (FC) map was an early attempt to present the functional connections between physiological units in the context of the anatomical connectivity derived from SCKAN. This map is shown below for reference. Extensive user research encouraged the exploration of alternative approaches to generating these kinds of functional connectivity maps. FC maps have now been redefined to describe specific physiological relationships (often within an anatomical context) and are published as a dataset on the SPARC Portal. These new FC maps can be accessed through the Data & Models page and documentation on their discovery and use is available. The number of new FC maps will continue to increase.
Updated 3 months ago