Flatmap Sources
The flatmap source files must follow a particular structure in order to successfully generate maps with no issues. A clone repository of the rat flatmap on the SPARC Portal has been set up on GitHub.
SVG File
The SVG file of the flatmap sources represents the base drawing of the flatmap. As can be seen in the rat_flatmap.svg file, each shape has a unique identifier and a closed shape. The nomenclature of the features in the SVG is important as it must be consistent with the JSON files. Below is a table of identifiers for types of features in the SVG.
| Feature | Identifier |
|---|---|
| Closed shape | .id(FEATURE_ID) |
| Centreline | .centreline id(CENTRELINE_ID) |
Table 1: The respective identifiers for features in the SVG file. These IDs must be strictly followed as the JSON files utilize them.
JSON Files
The JSON files of the flatmap sources contain information on the visual aspects of the SVG file. Here, knowledge on what the shapes in the SVG represent and the manner of their connections are laid out in the form of annotations. The identifiers used must match those in the SVG, and the layout of the files are very specific. For more information see the docs section of mapmaker.
Anatomical_map.json
The anatomical_map.json contains classes of features and their relative ontological ID - typically UBERON or ILX, as per the SciCrunch database. The layout is typically as follows:
{
“CLASS_ID”: {
“term”: “ONT_TERM”,
“name”: “FEATURE_NAME”
}
}Figure 1:
Properties.json
The properties.json file contains information on what features belong to which classes, as well as the connectivity knowledge between features. The structure can be split into two sections - the features definitions and the networks definitions.
Features in properties.json
The features section layout is typically as follows:
{
“features”: {
“FEATURE_ID”: {
“class”: “CLASS_ID”
}
}Figure 2:
The feature_ID must be consistent with the ID used in the SVG file, for the relative feature(s). The class_ID must be consistent with the relative class that has been set up in anatomical_map.json.
Networks in properties.json
The networks section layout is typically as follows:
{
“networks”: [
{
“centrelines”: [
{
“id”: “CENTRELINE_ID”,
“models”: “NERVE_ONT_TERM”
“connects”: [
“FEATURE_ID1”,
“FEATURE_ID2”
],
“contained-in”: [
“FEATURE_ID3”
]
}
}
}Figure 3:
The fields in the networks section are explained in Table 2. As with the anatomical_map.json, the identifiers used must be consistent with those in the SVG file.
| Required fields | |
| id | A unique identifier specific to what the centreline represents. |
| connects | A list of closed shapes that the respective centreline connects. A minimum of two features is required as centrelines must have a start and end point. |
| Optional fields | |
| models | A unique identifier for the anatomical structure that the centrelines represents. Must only be used for centrelines not divided into segments. |
| contained-in | A list of close shapes that the respective centrelines passes through. These must be unique and specific to the respective centreline (i.e. if more than one centreline passes through a shape it is of no use to list the shape in the contained-in field). |
Table 2: The required and optional fields for the structure of centrelines in the properties.json file.
Manifest.json
The manifest.json ties together the other JSON files and the SVG file. The structure of the manifest is typically as follows:
{
“id”:”FLATMAP_NAME”,
“models”: “FLATMAP_ONT_ID”,
"description”: “description.json”,
“properties”: “properties.json”,
“sckan-version”: “production”,
“sources”: [
{
“id”: “FLATMAP_ID”,
“href”: “FLATMAP_SVG”,
“kind”: “base”
}
],
“anatomicalMap”: “anatomical_map.json”,
“connectivityTerms”: “connectivity_terms.json”,
“neuronConnectivity”: [
“https://apinatomy.org/uris/models/ard-arm-cardiac”,
“https://apinatomy.org/uris/models/bolser-lewis”,
“https://apinatomy.org/uris/models/bronchomotor”,
“https://apinatomy.org/uris/models/keast-bladder”,
“https://apinatomy.org/uris/models/pancreas”,
“https://apinatomy.org/uris/models/sawg-distal-colon”,
“https://apinatomy.org/uris/models/sawg-stomach”,
“https://apinatomy.org/uris/models/spleen”
]
}Figure 4:
Given these files are set up in the correct manner, the desired flatmap should be able to generate with no issues.
Examples
As can be seen in Figure 5, from the anatomical_map.json file of the rat_flatmap files, the class urinary_2 represents the urinary bladder of the body.
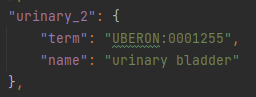
Figure 5: Extracted from the anatomical_map.json file, the urinary_2 class represents the urinary bladder and has the appropriate term associated
The features in the SVG that represent the urinary_2 class are defined in the properties.json (Figure 6).
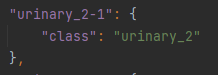
Figure 6: Extracted from the properties.json file, the urinary_2-1 belongs to the urinary_2 class. In the SVG file, the feature that represents the urinary bladder will be labelled with ‘urinary_2-1’
Note that the use of “-1” in the feature ID allows multiple features to belong to the same class (i.e. in Figure 7, urinary_7-1 and urinary_7-2 both belong to the urinary_7 class).
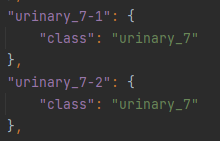
Figure 7: Extracted from the properties.json file, the eatures with IDs ‘urinary_7-1’ and ‘urinary_7-2’ belong to the class ‘urinary_7’. This correlates to two separate features in the SVG with IDs ‘urinary_7-1’ and ‘urinary_7-2’.
Within the networks section of the properties.json, urinary_13-1 is referenced as the endpoint of the centreline n_58 (Figure 8).
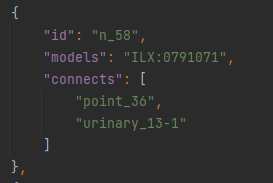
Figure 8: Extracted from the properties.json file, the centreline ‘n_58’ connects features ‘point_36’ and ‘urinary_13-1’. It also represents a nerve, therefore has an ontological ID associated with it.
Updated 8 months ago