Formatting SPARC Datasets
The SPARC dataset structure (SDS) is very flexible and support many different types of experiments. Use this to help you determine how to structure yours.
Organization of a SPARC Dataset
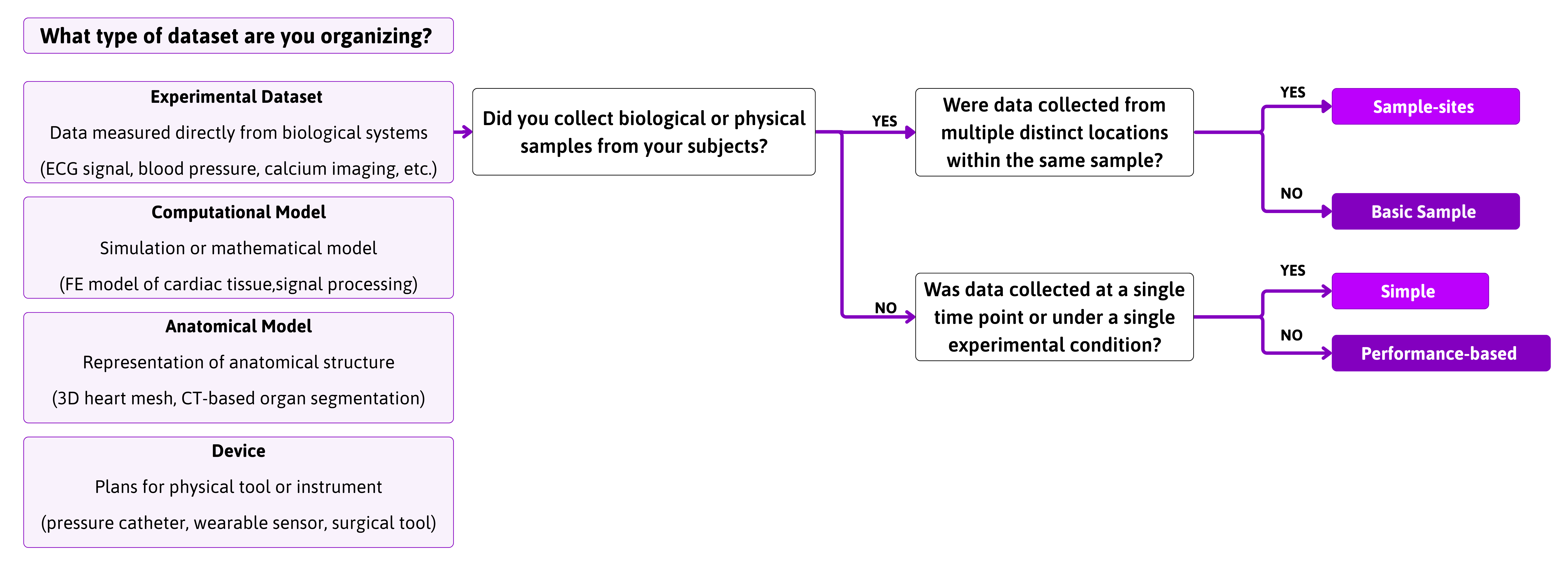
SPARC data is organized into datasets, each containing many files. While all use the SDS, what aspects of it will be used, e.g. how your data or models are formatted will vary depending on the structure of the experiment and how it is collected.
Note: The Pennsieve platform underlying SPARC allows one to publish code Datasets, these Pennsieve code datasets can be referenced in SPARC daatasets.
Coming soon: examples of how to structure certain types of experiments.
Updated 4 months ago