SDS: Structured Folders and Metadata Files
Overview
SPARC dataset includes a folder structure that follows the SDS convention along with metadata contained in a set of spreadsheet templates. These mandatory descriptive files containing the metadata are essential for understanding your dataset. Adding information about your experiment (metadata) and formatting your data properly aids findability and data reuse (both human and machine).
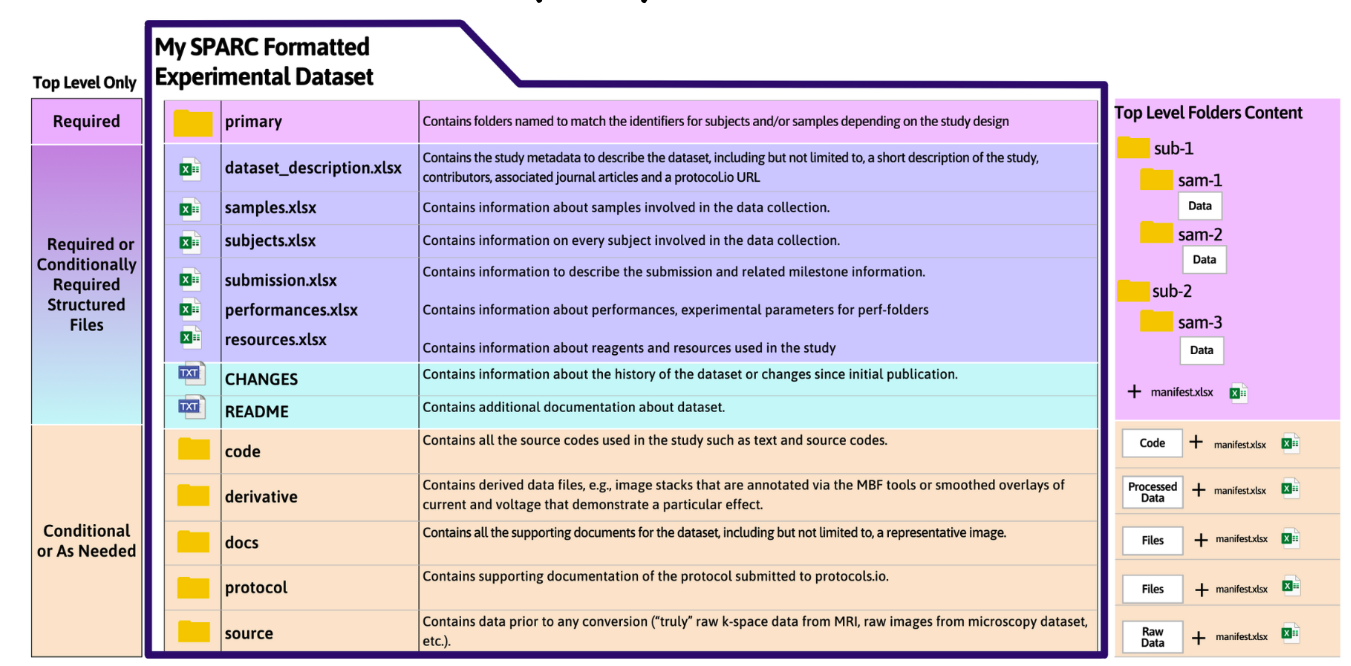
Example of dataset organized according to SDS Version 2.1

Example of a dataset organized locally according to the SPARC Dataset Structure
Pennsieve treats these files as data. Therefore, any changes to them after the dataset has been published will result in a re-publication and updated DOI. Learn more about the Changes to Published Datasets Policy
Continue reading to learn more about the Folders and Metadata Files.
Data Folders
All data files are organized into top-level (dataset-level) folders. Some of these are required, and some are optional, dependent upon the type of data you want to include in your dataset.
Sets of metadata and descriptive files may also be required in these folders, as described above.
Data files are organized into 3 different top-level folders, depending on the type of data:
- primary
- source
- derivative
Refer to Naming Requirements for the standardized prefixes for each type of data folder.
Primary Folder
This folder is required and contains all of your experimental data.
primary: a required folder for experimental data. This dataset-dependent folder contains all folders and files for experimental subjects and/or samples (e.g., time series data, tabular data, clinical imaging data, genomic, metabolomic, or microscopy data). The data generally have been minimally processed so they are in a form ready for analysis.
Within the primary folder, data is organized by subjects or samples. All subjects and samples will have a unique folder with a standardized name corresponding to the exact names or IDs as referenced in the subjects and samples metadata file. The structure of this folder must match the subjects, samples, and pools described in the top level metadata files (e.g., subjects.xlsx, and samples.xlsx).
- Examples may include time-series data, tabular data, clinical imaging data, genomic, metabolomic, microscopy data.
- The data generally have been minimally processed so they are in a form ready for analysis.
Data is organized by subject/sample, and all subjects and samples will have a unique folder within the Primary Folder.
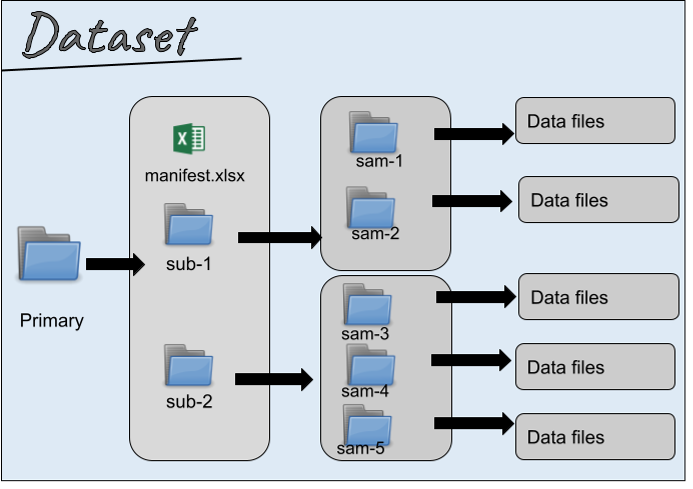
Generalized diagram of the SDS
Source Folder
This folder is only required if unaltered, raw files from an experiment are included in the data. If so, they will be placed in this folder.
- An example would be the “truly” raw k-space data for a Magnetic Resonance (MR) image that has not yet been reconstructed.
- In this case, the reconstructed DICOM or NIFTI files would be found within the primary folder
Please note, there are no specific requirements about folder naming inside this Source Folder. However, we recommend using the sub-, sam-, perf- structure similar to the primary folder’s naming conventions. Refer toNaming Convention section.
Derivative Folder
This folder is only required if derivative data exists. If so, they will be placed in this folder.
- derivative: required folder, if derivative data exists. This folder contains data derived from the data in the primary data folder. For example, processed image stacks that are annotated via the MicroBrightField (MBF Biosciences) tools, segmentation files, or smoothed overlays of current and voltage that demonstrate a particular effect. Derived data should be organized into subject and sample folders, using the subject and sample IDs as the folder names, as with the primary data.
- Examples may include processed image stacks that are annotated via the MicroBrightField (MBF Biosciences) tools, segmentation files, or smoothed overlays of current and voltage that demonstrate a particular effect.
- If files are converted into a format other than what was submitted, these files are included in the derivative folder.
- Derived data should be organized into subject and sample folders, using the same subject and sample IDs as the folder names within the Primary Data folder.
Dataset Specific Folders and Files
Some metadata and descriptive folders and files are only required if your dataset includes an experimental approach or datatype that requires more information:
Subjects and/or Samples
If the dataset contains subjects, samples, and/or multiple data collection time points, the following files are required:
- subjects (xlsx, csv or json): Conditionally Required if subjects are used in the experiment producing the dataset. Contains updated fields with required and optional metadata fields providing information about subjects (model organism or animals) involved in data collection. Each subject and/or pooled subjects must be assigned a unique ID. This unique ID is used to name the data folders for individual subjects. For proper mapping of the data, folders containing experimental data need to exactly match the subject ID. All subject identifiers must be unique within a dataset and not contain any sensitive, identifiable information (for human subjects). Having each lab use consistent, unique subject identifiers across datasets is highly desirable to aid in connecting multiple experiments using the same subjects.
- samples (xlsx, csv or json): Conditionally Required if measurements are obtained from samples, e.g., tissue slices, derived from individual or pooled subjects. This file contains information about samples used to generate the data. Investigators must provide a unique ID for each sample which will be used to name the data folders, and the sample ID must match the folder ID exactly. Sample IDs must be unique within a single dataset. Each sample should also reference a subject from the subject file; a single subject (a research animal/donor) may be linked to multiple biological samples derived from that subject. If the samples are pooled from multiple subjects, the complete provenance must be specified in the subject file. The metadata present in the samples file should also explicitly note whether a sample was collected directly or was derived from another sample.
- performances (xlsx, csv or json): Conditionally Required if data were gathered from multiple distinct performances of one type of experimental protocol on the same subject or same sample (i.e. multiple visits, runs, sessions, or execution).
- sites (xlsx, csv or json): Conditionally Required if sharing experimental locations. It contains specific anatomical or experimental locations related to subjects or specimens. These could include locations like electrode placements, biopsy sites, or other defined locations.
If the dataset contains subjects or samples but the folder structure does not include subject and/or sample ids, the manifest (xlsx, csv or json) file is conditionally required. It will list all files and folders in the dataset, mapping them to specific entities like subjects and samples. This file helps ensure proper organization and provides key metadata for each file. Learn more about the manifest file.
Code
These files and folder are only required if code is used in the generation of the data. If so, the folder contains all the source codes used in the study. Please note, if your code is on GitHub, you don’t need to share it here. Simply put the link in your “Dataset_description” file in the related identifier field.
Examples may include text and source code (e.g. MATLAB, etc.), script to plot the results, script to open and read raw data files, source code for computational models.
Links to supporting code that provides added value to the dataset can be included in the metadata description but does not have to be uploaded here.
- code (folder): Conditionally required a required folder only if code is used in the generation of the data; the folder contains all the source codes used in the study such as text and source code (e.g., MATLAB, etc.). Links to supporting code that provides added value to the dataset can be included in the metadata description but does not have to be uploaded here.
- code_description (xlsx, csv or json): Conditionally required if submitting code or a computational model. When submitting code, it documents any code or software included with the dataset. When submitting a computational model, it contains information to describe the code in terms of its quality. Code RRIDs and ontologies are required.
- code_parameters (xlsx, csv or json): Optional file containing information describing specific parameters to run a code. This file can be found in older versions of SDS.
Supplemental Folders
Protocol Folder
This folder is an optional folder that contains supplementary files to accompany the experimental protocols submitted to Protocols.io.
IMPORTANT: This is not a substitution for the experimental protocols which are required to be created and shared with SPARC or RE-JOIN workspace on Protocols.io.
Documents Folder
This folder is optional and contains all the supporting documents for the dataset. An example would be a representative image for the dataset. Unlike the readme file, which is a text document, docs can contain documents in multiple formats, including images.
Structured Metadata Files
Dataset-level files
Dataset Description File
dataset_description (xlsx, csv or json) Required-all datasets must contain a dataset_description file. provides study metadata describing the dataset. An example image is below.
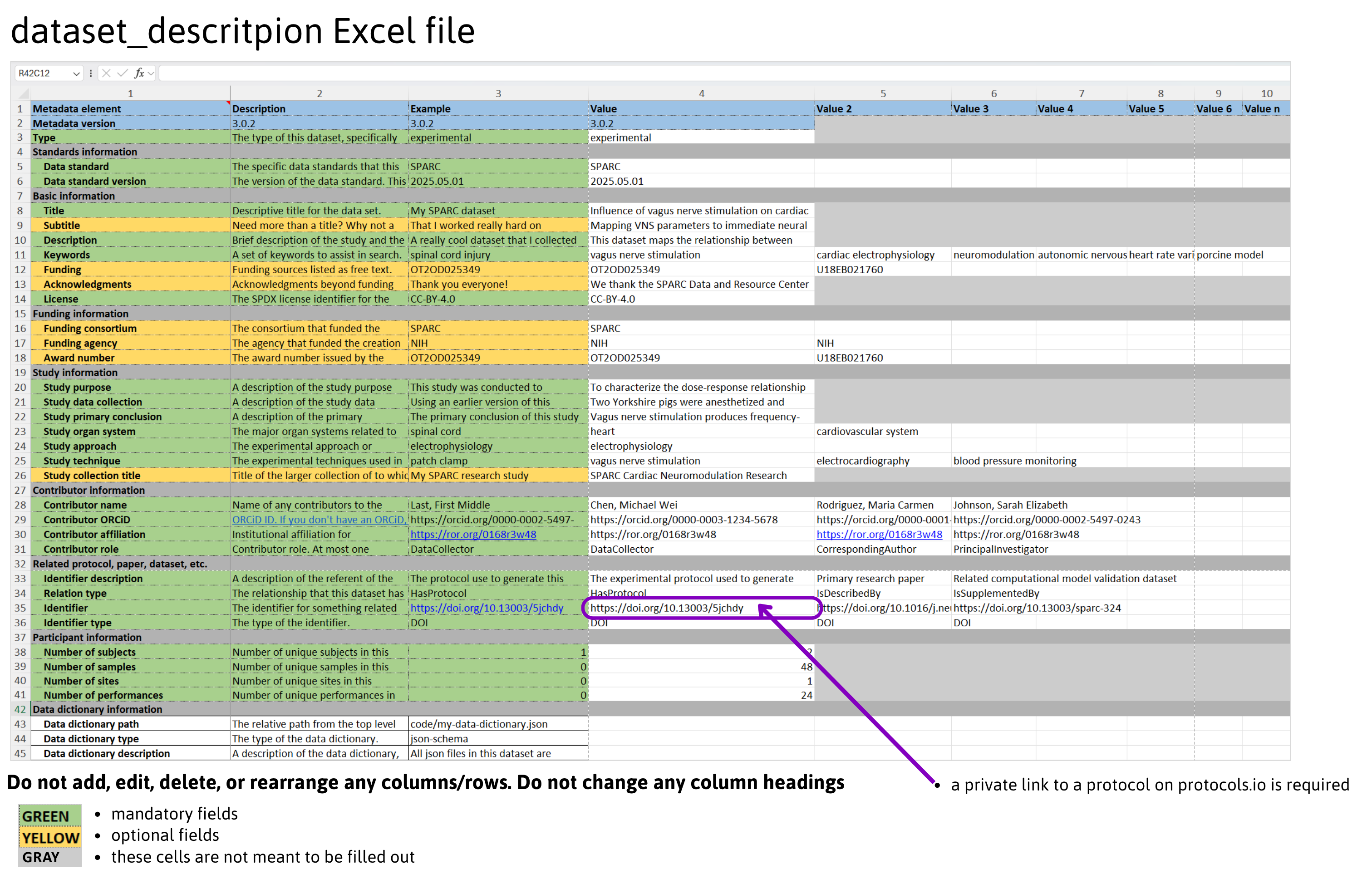
The dataset_description.xlsx file includes fields called Metadata version & Standards Information that specify the metadata version. These fields are not to be changed by data submitters, as it is used to properly align different metadata releases. The dataset_description file also includes sections for researchers to describe their dataset.
- Basic information about the dataset like title, keywords
- Funding information consortium, agency # award #
- Study information with parts similar to a structured abstract like study purpose, data collected, conclusions, organ system, approach (modality), and techniques. If the dataset is part of a larger study, this is also where we link it to other datasets.
- Contributor information names, ORCIDs, affiliation & role
- Related protocol, paper, dataset, etc. area where these identifiers are shared
- Participant information number of subjects, sites and performances
- Data dictionary information information about associated data dictionary
README File
README (txt): Required file provided by Investigators that contains necessary details for reuse of the data, beyond that which is captured in structured metadata. Some information that should be included are:
- How would a user use the files that are provided? e.g., "first open file X and then look at file Y."
- What additional details do users need to know? Are some subjects missing data?
- Are there warnings about how to use the data or code?
- Are there appropriate/inappropriate uses for this data?
- Are there other places that users can go for more information? e.g., did you provide a GitHub repository, or are there additional papers beyond what was provided in the metadata form?
Resources File
resources (xlsx, csv or json): Optional file containing information to describe resources used in the experiments (RRID, URL, vendor, version, additional metadata).
Changes File
CHANGES (txt): Conditionally Required file to document any changes made to a datasets after it has been published.
Manifest Files
Within the top-level (dataset-level) Data Folders described above, SDS requires that manifest spreadsheets that provide information about specific files within a folder. For example, a folder may contain a file named mov_colon_stim.mp4. To make sure that your data are understandable, provide a brief description of the contents of this file along with other key metadata.
File-level manifest(xlsx, csv or json) are required in all top-level folders (listed above). It is also acceptable to have a single manifest file located at the root of the folder tree.
Further details about the structure are provided in this presentation and the project’s white paper.
Manifest Files are best created AFTER your top-level folders are organized, as they describe the contents of the folder.
Please note that if you’re using SODA to prepare and upload your dataset, the program automatically creates a list of files for the manifest spreadsheet and prompt you to add a description of each file in a systematic way during the uploading process. If you are uploading datasets without SODA, you will need to create this file yourself for each submitted top-level folder.
Best Practices
- Make sure you are always using the most recent SDS template version.
- Some of the folders and metadata templates have specific file names. The names of these key folders and the templates must be kept the same between different dataset submissions.
- Do not add, edit, or delete required columns/rows or change any column headings in any mandatory descriptive files (templates).
- If you wish to add additional metadata (always encouraged), please append a new column to the right-hand side (Subject and Sample files) to include critical metadata not corresponding to available columns. The dataset_description.xls template should not be changed at all.
- Leave fields empty when there is no information available at the time of submission.
- Using SODA greatly simplifies and automates creating the required manifest files
Updated 9 months ago