Map-Tools
Overview
Map-tools is a jupyter notebook package that provides information on individual neuron connections, according to the knowledge in SCKAN. It can be used to understand each connection and the respective anatomical features involved.
Source Code
The Map-Tools source code is hosted on GitHub.
Installation
Map-tools is best run under a python environment and is reliant on Poetry. Map-tools can then be installed by cloning the GitHub repository.
$ git clone https://github.com/AnatomicMaps/map-toolsHaving installed Poetry via the official installer instructions, clone and enter the map-tools directory. Install a Poetry environment with
$ poetry installUsage
Once installed, map-tools can be run by opening Jupyter Notebook. In the map-tools directory, run jupyter notebook via poetry.
$ poetry run jupyter notebookThe command will provide access to the notebook with a URL, which should then be pasted into a browser window (Figure 1).
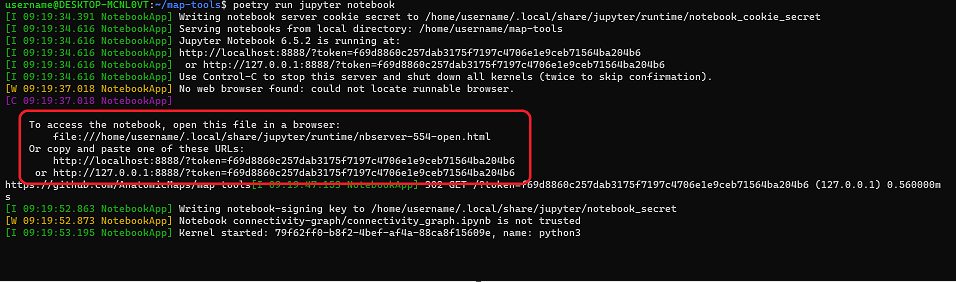
Figure 1: Screen capture of successful activation of the map-tools Jupyter Notebook. Follow the instructions outlined by the red box to access the notebook.
Once opened, navigate to and open the connectivity_graph.ipynb notebook in the connectivity-graph folder. A valid SCICRUNCH Key must be input in order to draw knowledge from SCKAN. This can be done by inserting the key under a scicrunch_key field as shown in Figure 2. Note that a SciCrunch key can be generated by setting up a SciCrunch account.
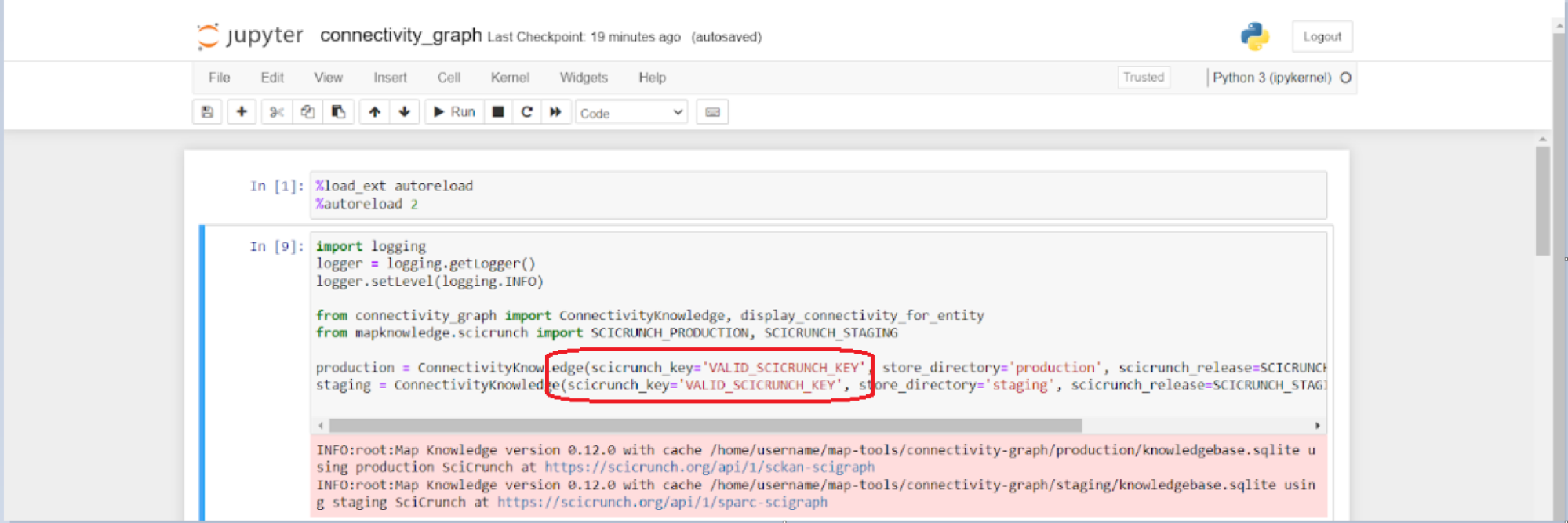
Figure 2: Screen capture of the connectivity_graph.ipynb file. Inset a valid SciCrunch API Key under a ‘scicrunch_key’ field as outlined by a red box.
Neuron paths from ApiNATOMY Models can now be visualized, as seen in Figure 3. The desired neuron path can be visualized by changing the entity field. Table 1 represents the ilxtr attribute for each model. Note that the ID of each path is dependent on the visualisation of paths in each model.
| Model | ilxtr (‘X’ represents path ID) |
|---|---|
| ApiNATOMY model of bronchomotor control | neuron-type-bromo-X |
| ApiNATOMY model of the spleen | neuron-type-splen-X |
| ApiNATOMY model of the stomach | neuron-type-sstom-X |
| Ardell-Armour model of the heart | neuron-type-aacar-X |
| Bolser-Lewis model of defensive breathing | neuron-type-bolew-unbranched-X |
| Keast model of the bladder | neuron-type-keast-X |
| SAWG model of the distal colon | neuron-type-sdcol-X |
Table 1: ilxtr attributes of each path in the respective ApiNATOMY models. These are used to visualise said paths in the connectivity_graphy.ipynb file.
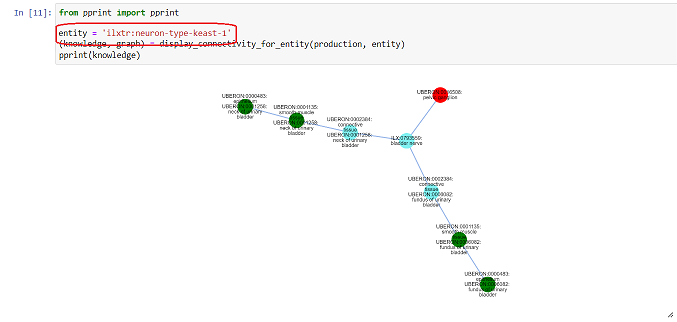
Figure 3: Visualization of the neuron path 1 from the Keast ApiNATOMY model. By changing the ‘entity’ attribute, the various different neuron paths can be explored.
Updated 8 months ago